QC Methods Showdown: QClus vs SoupX vs Doublet Detection in Human Atrial Data
Introduction
Quality control (QC) is a critical step in single-cell and single-nucleus RNA sequencing (snRNA-seq) analysis. Poor quality control can introduce technical artifacts, ambient RNA contamination, and doublets that may confound biological interpretation. In this post, I compare different QC strategies applied to human atrial snRNA-seq data to evaluate their impact on downstream analysis.
QC Methods Overview
QClus [1] is a clustering-based quality control method that identifies low-quality cells by detecting outlier clusters in the quality metric space. It uses unsupervised learning to distinguish technical artifacts from biological heterogeneity without requiring hard thresholds.
SoupX [2] estimates and removes ambient RNA contamination in droplet-based single-cell RNA-seq data. It models the “soup” of cell-free mRNA molecules present in the droplet solution that can be erroneously captured in cell barcodes, particularly affecting marker gene expression patterns.
Scrublet [3] identifies doublets by simulating artificial doublets from the data and comparing them to observed transcriptomes. Doublets occur when two cells are captured in the same droplet, creating artificial cell states that can mislead clustering and cell type identification.
Study Objective
The goal of this comparison is to evaluate how different QC strategies affect:
- Cell filtering decisions
- Clustering and cell type identification
- Differential gene expression analysis
I wanted to understand whether more aggressive QC methods provide meaningful improvements or if they risk removing genuine biological signals.
Dataset
For this analysis, I used the dataset from GSE255612 [4], published by Hill et al. [5]. This dataset contains single-nucleus RNA-seq data from 16 healthy controls and 18 atrial fibrillation patients, providing a robust foundation for comparing QC methods in cardiac tissue analysis.
Initial Data Assessment
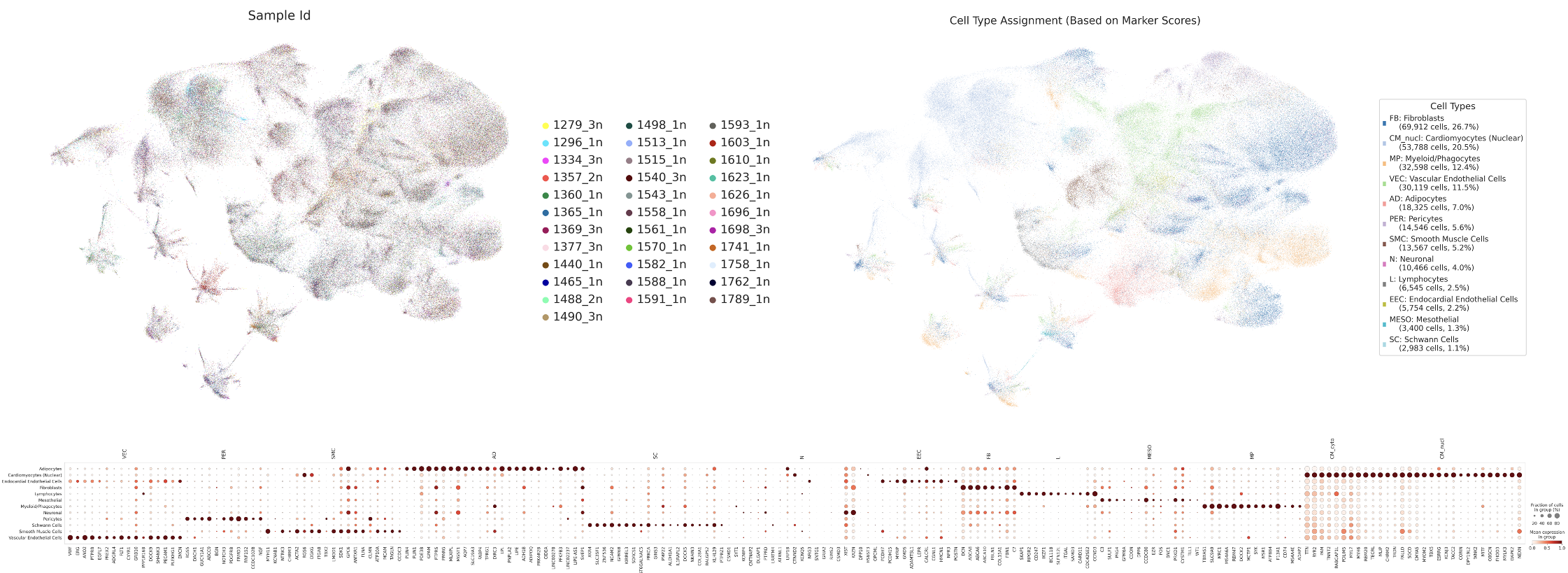
The data were first integrated across samples using Harmony batch correction. This initial visualization shows the raw, unfiltered dataset with all nuclei included, serving as our baseline for comparison.
QC Metric Calculation
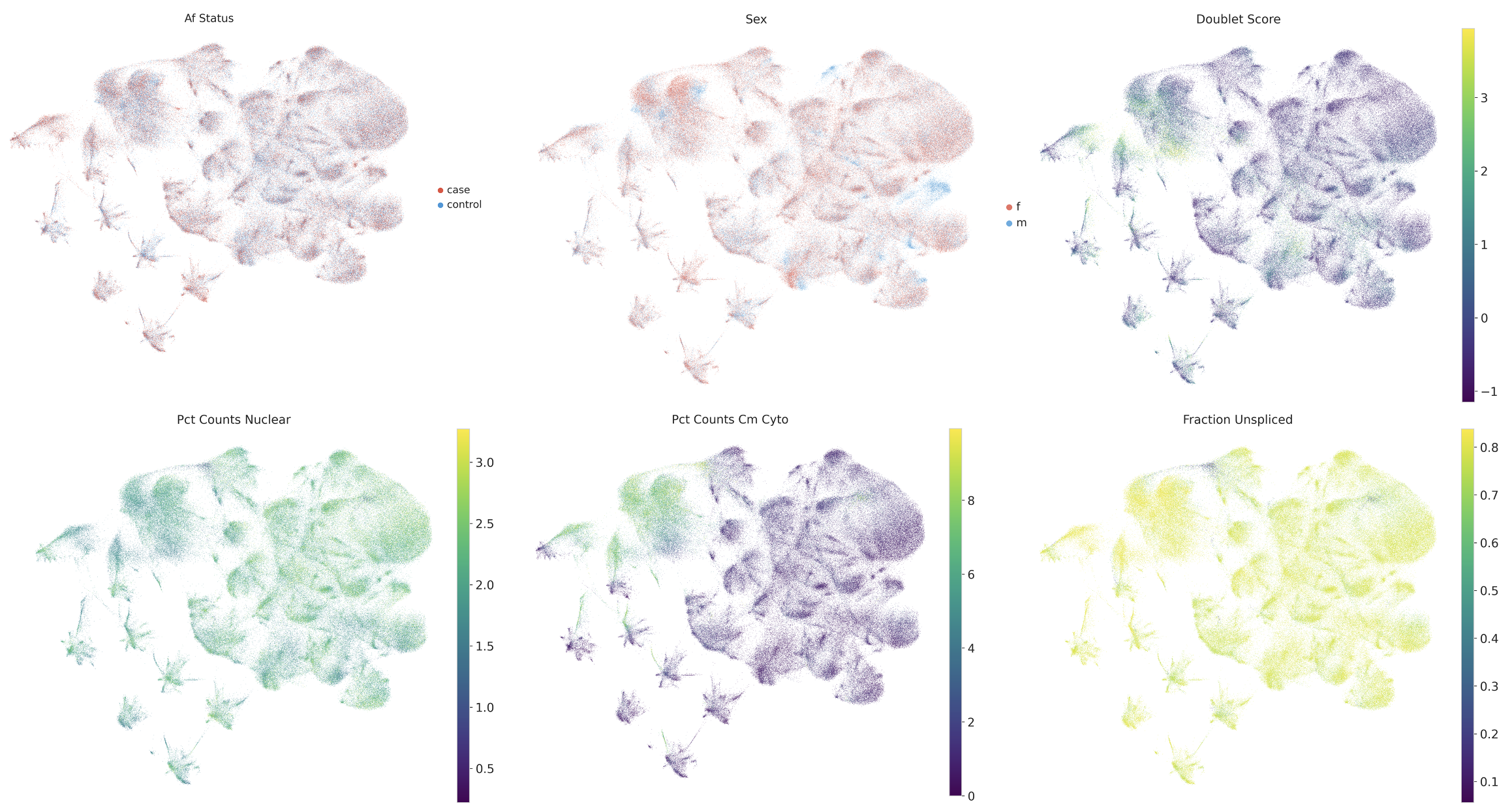
To assess nuclear quality and potential cytoplasmic contamination, I calculated two custom scores:
Nuclear Fraction Score based on expression of nuclear-enriched genes:
nuclear_genes = ['MALAT1', 'NEAT1', 'FTX', 'FOXP1', 'RBMS3', 'ZBTB20',
'LRMDA', 'PBX1', 'ITPR2', 'AUTS2', 'TTC28', 'BNC2',
'EXOC4', 'RORA', 'PRKG1', 'ARID1B', 'PARD3B', 'GPHN',
'N4BP2L2', 'PKHD1L1', 'EXOC6B', 'FBXL7', 'MED13L',
'TBC1D5', 'IMMP2L', 'SYNE1', 'RERE', 'MBD5', 'EXT1', 'WWOX']
Cardiomyocyte Cytoplasmic Score based on cardiomyocyte-specific genes:
cm_cyto_genes = ["TTN", "RYR2", "PAM", "TNNT2", "RABGAP1L",
"PDLIM5", "MYL7", "MYH6"]
These metrics help identify nuclei with abnormal RNA composition that may represent technical artifacts.
QC Method Comparison: Which Cells Are Filtered?
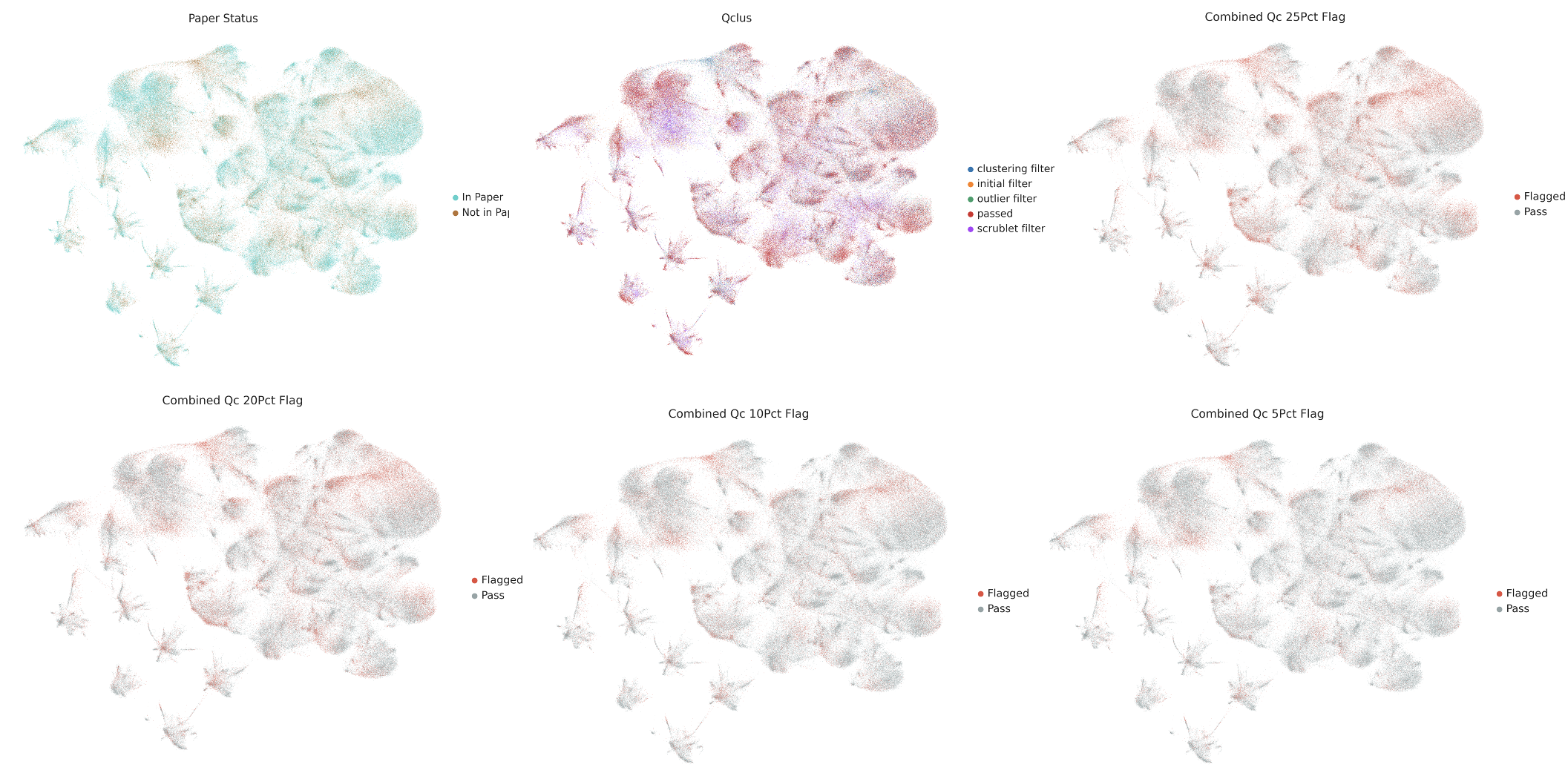
This visualization reveals the spatial distribution of cells targeted for removal by each method. Interestingly, different QC approaches identify distinct subpopulations as low-quality, with varying degrees of overlap. Some methods are more conservative while others apply more stringent filtering.
Quantifying QC Method Agreement
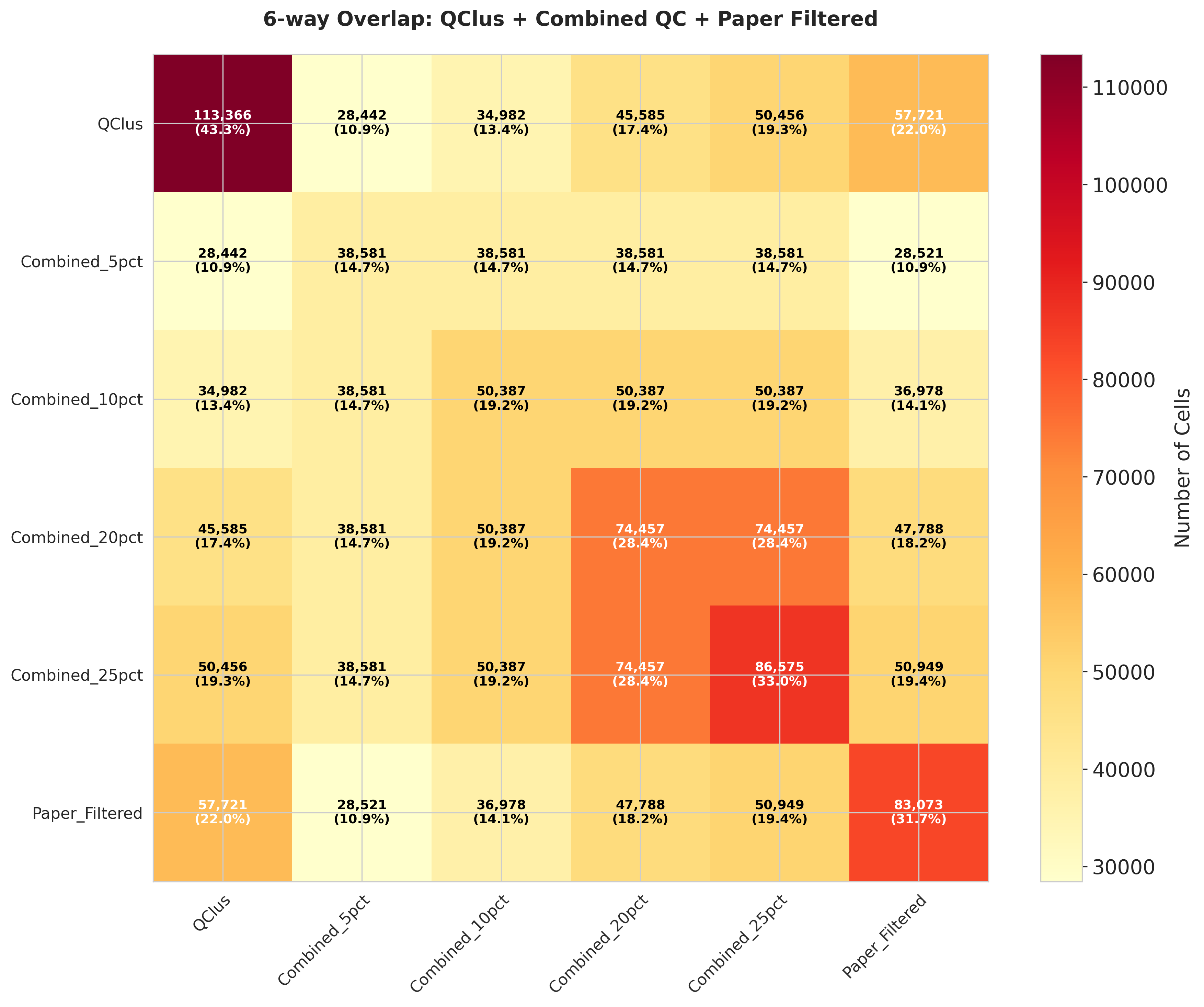
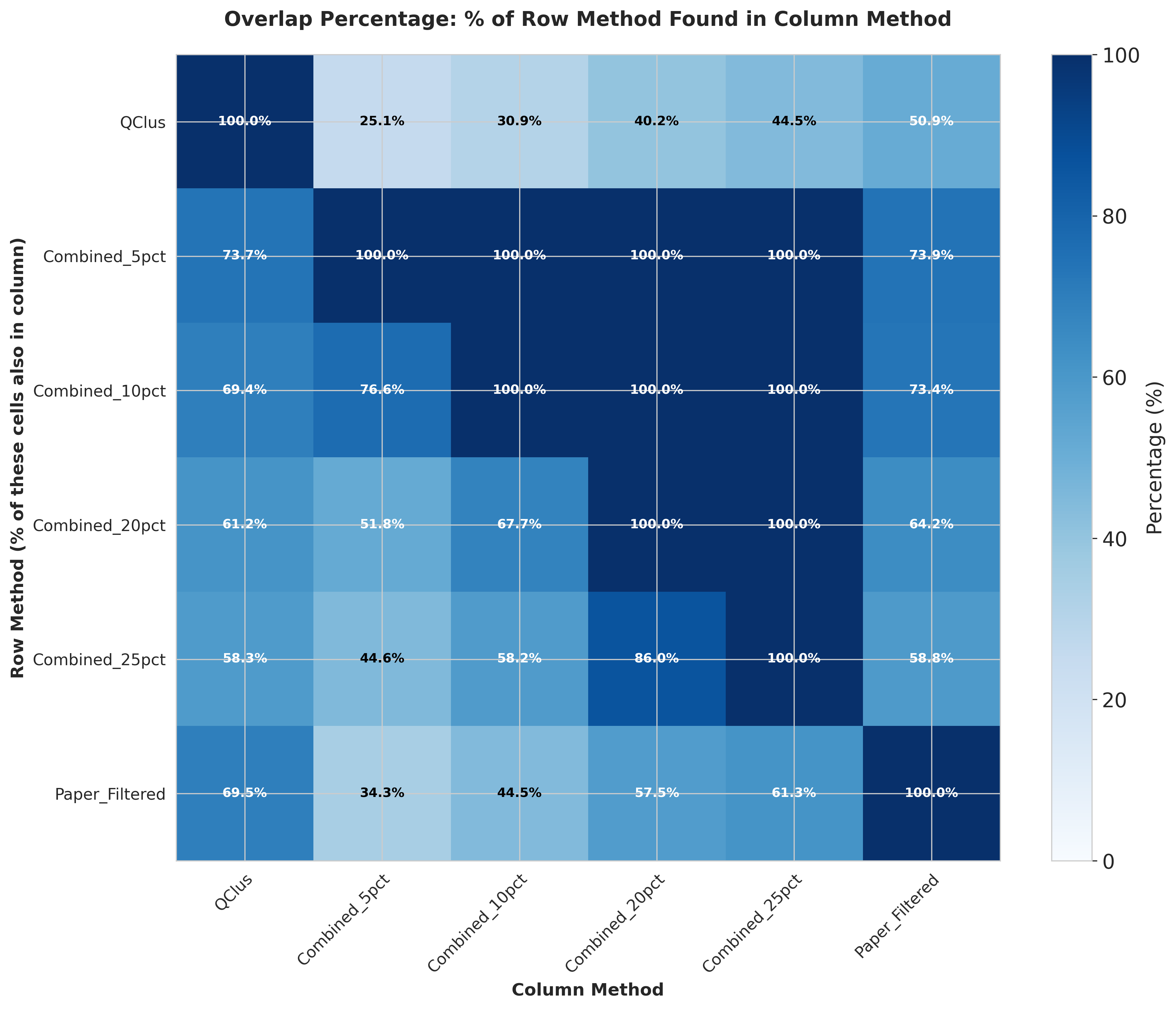
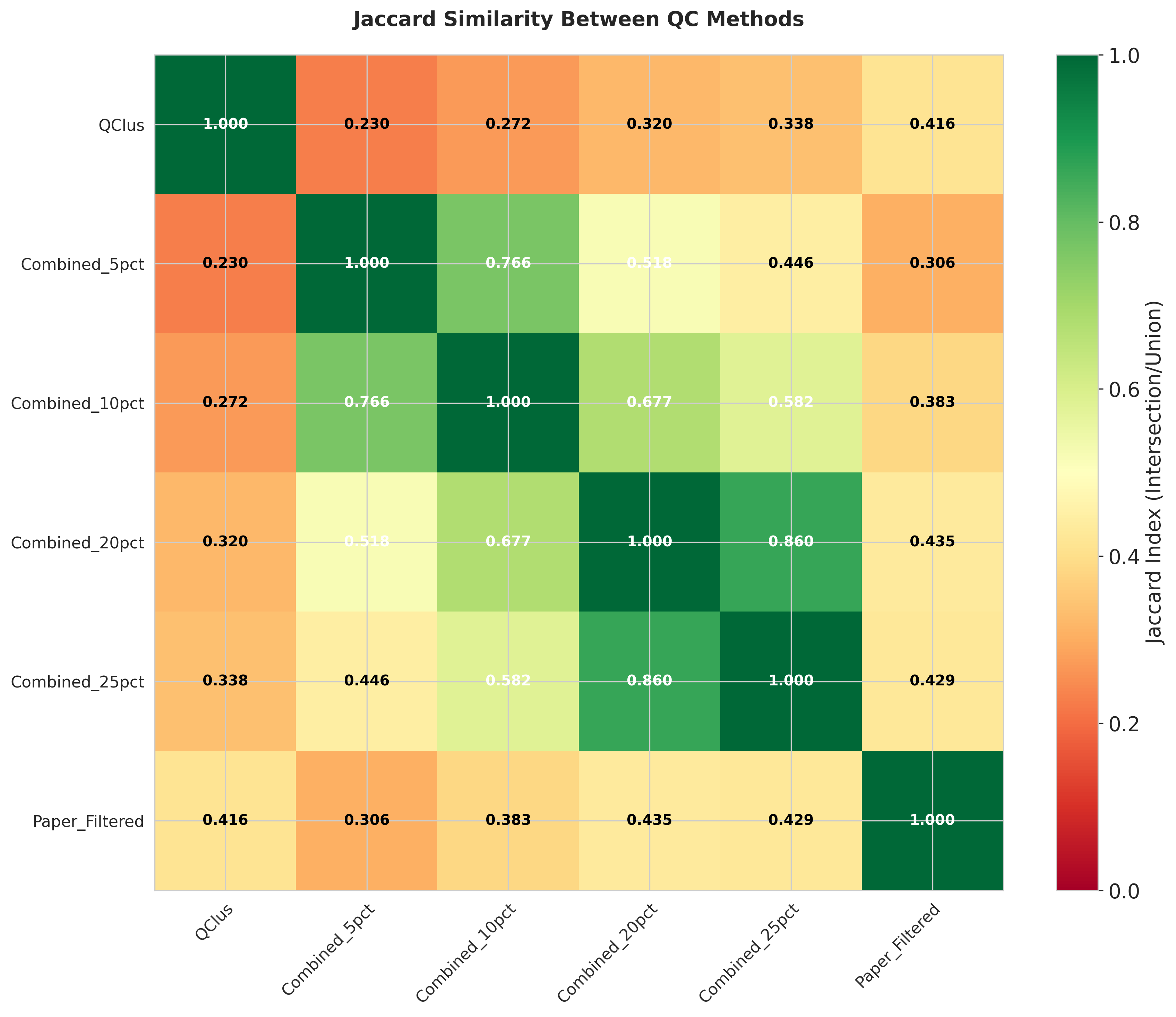
These heatmaps quantify the agreement between QC methods:
- Left: Total number of cells flagged for removal by each method
- Middle: Percentage of overlap in filtered cells between method pairs
- Right: Jaccard distance measuring dissimilarity between filtered cell sets
The analysis reveals substantial variation in both the number of cells filtered and the specific cells targeted by each method.
Impact on Clustering After Filtering
When we re-run dimensionality reduction and clustering after applying each QC filter, we can observe the structural changes in the data. Each method produces a slightly different landscape, though major cell populations remain largely consistent. The QSF condition represents cells that pass both QClus and SoupX (25% threshold) filters.
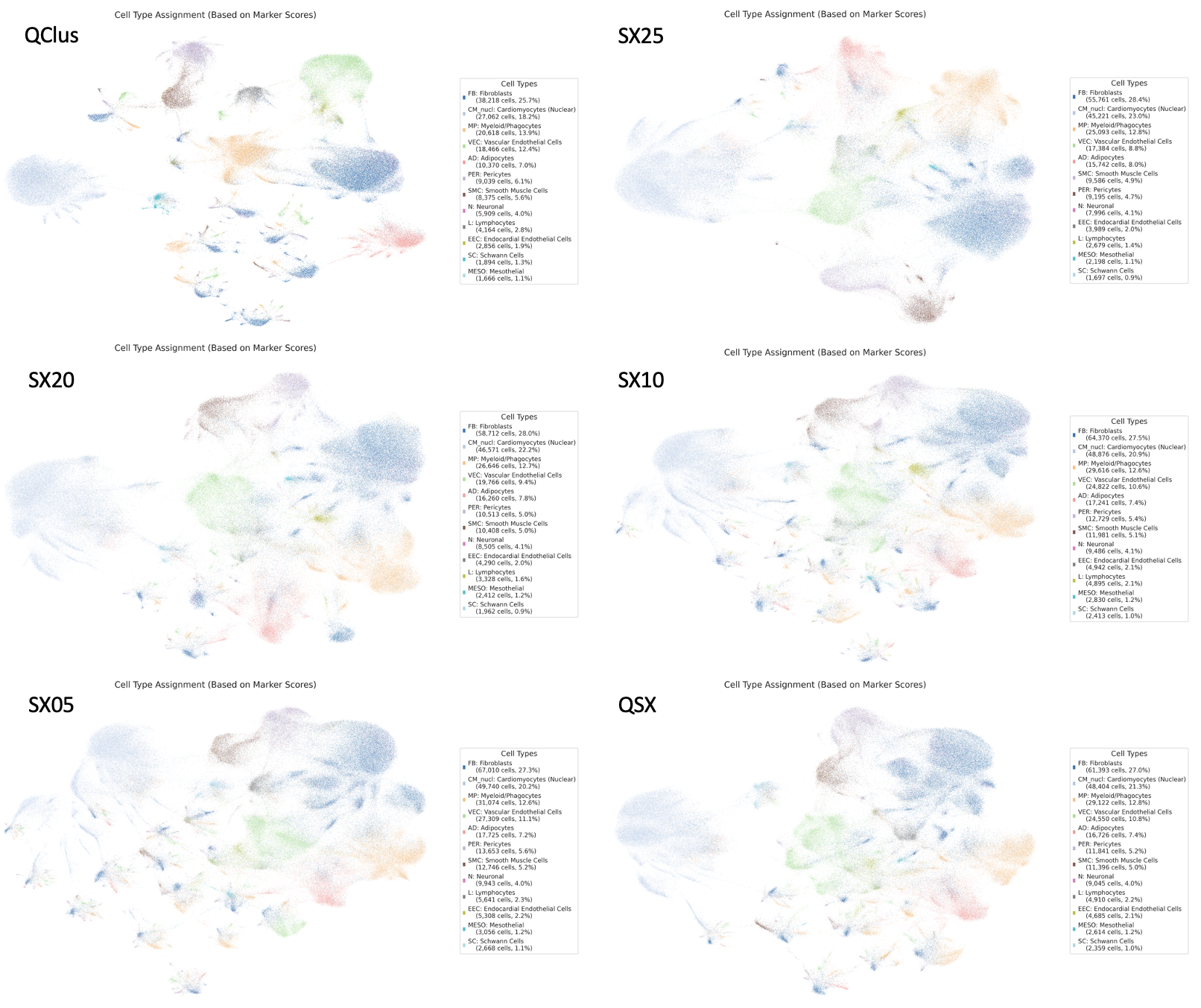
Cell Type Annotation Consistency
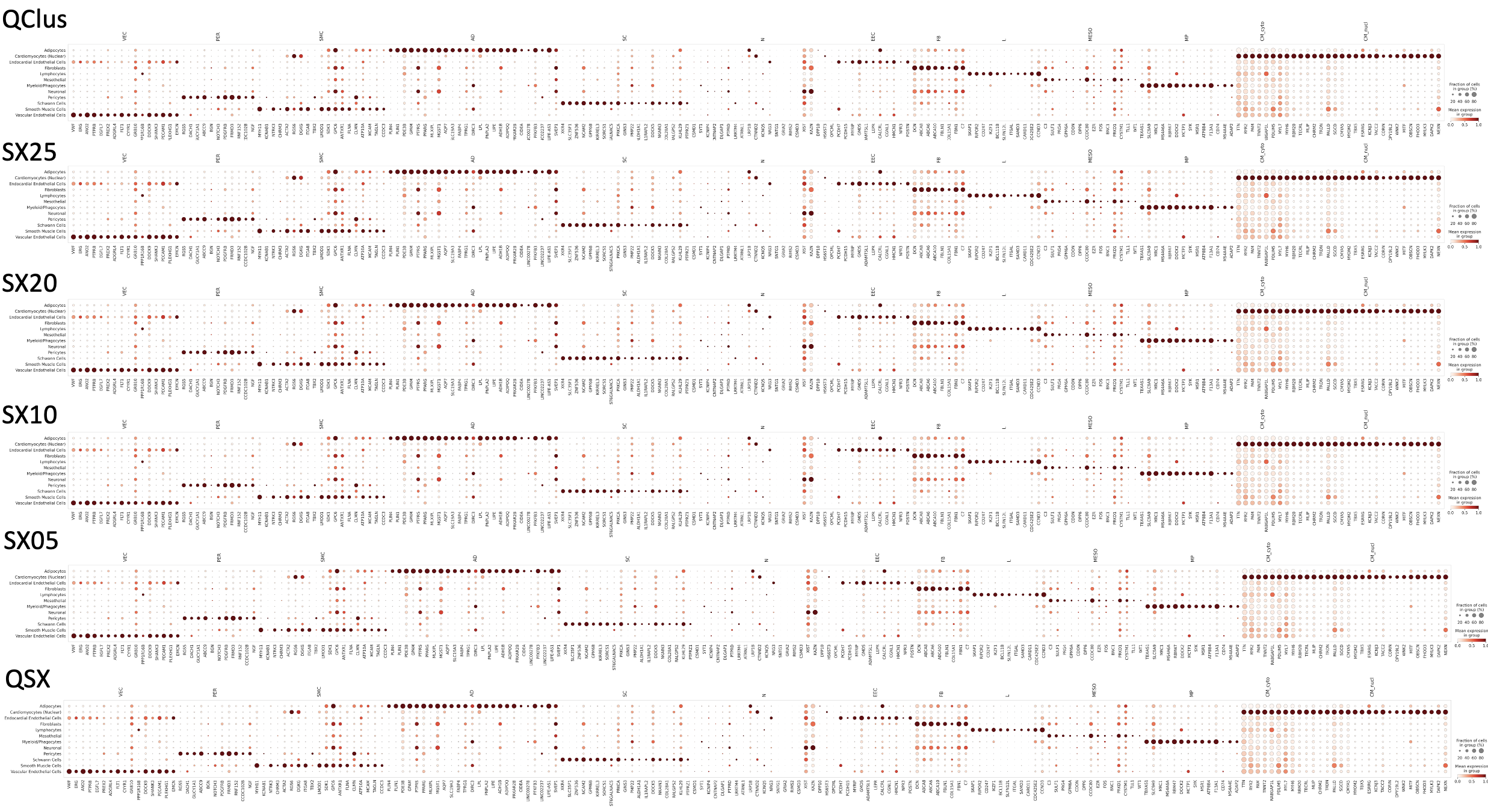
Despite differences in filtering strategies, cell type annotations remain remarkably consistent across QC methods. This suggests that the major cell type identities are robust to QC variation, and different QC approaches don’t dramatically alter our ability to identify known cardiac cell populations.
Impact on Differential Gene Expression
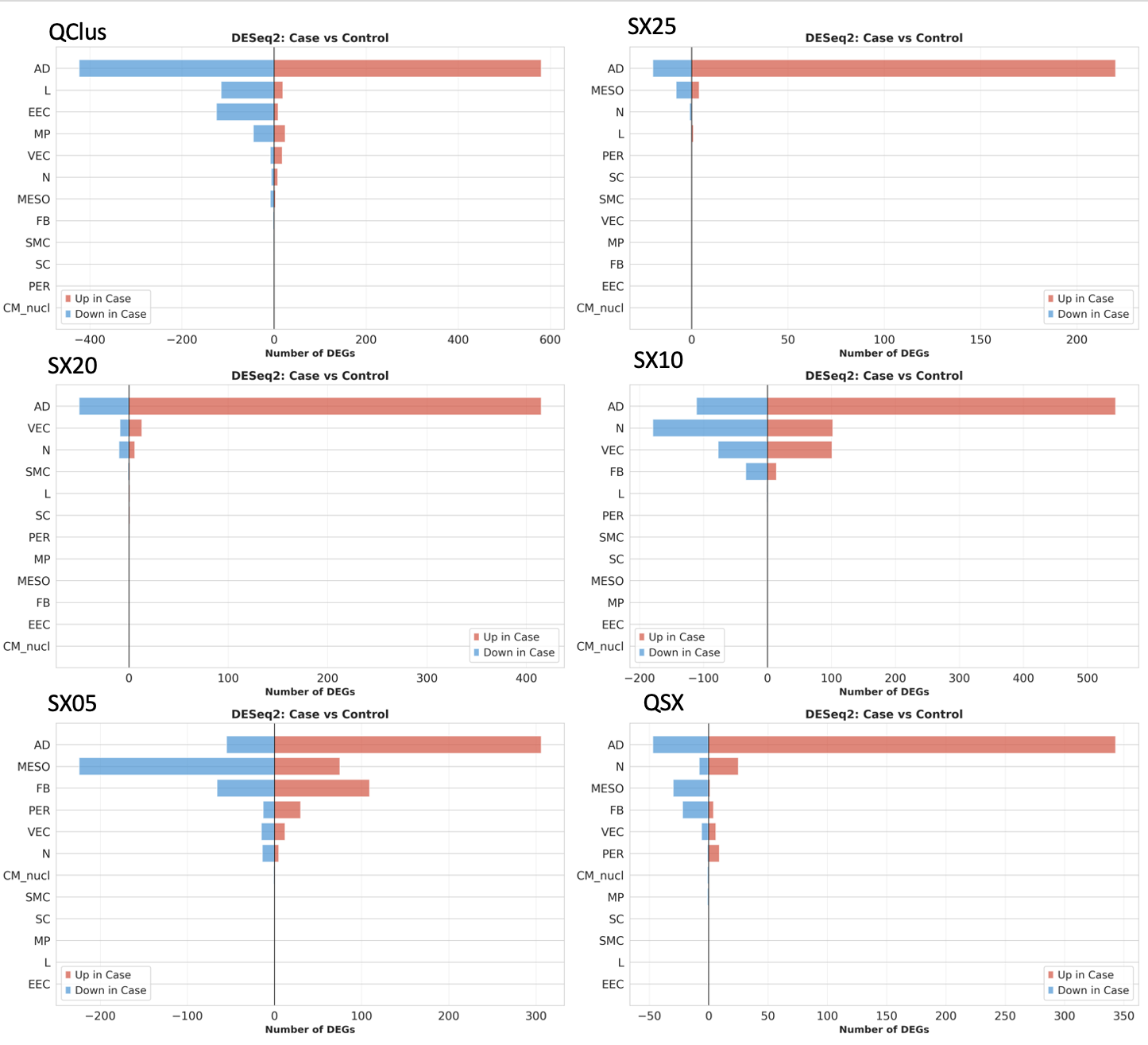
The most striking differences emerge in differential gene expression analysis. While the direction and magnitude of gene expression changes show consistent patterns across QC methods, the statistical significance varies considerably. This demonstrates that QC choices can substantially impact which differentially expressed genes (DEGs) reach significance thresholds, potentially affecting biological interpretation.
Top DEGs Across Cell Types
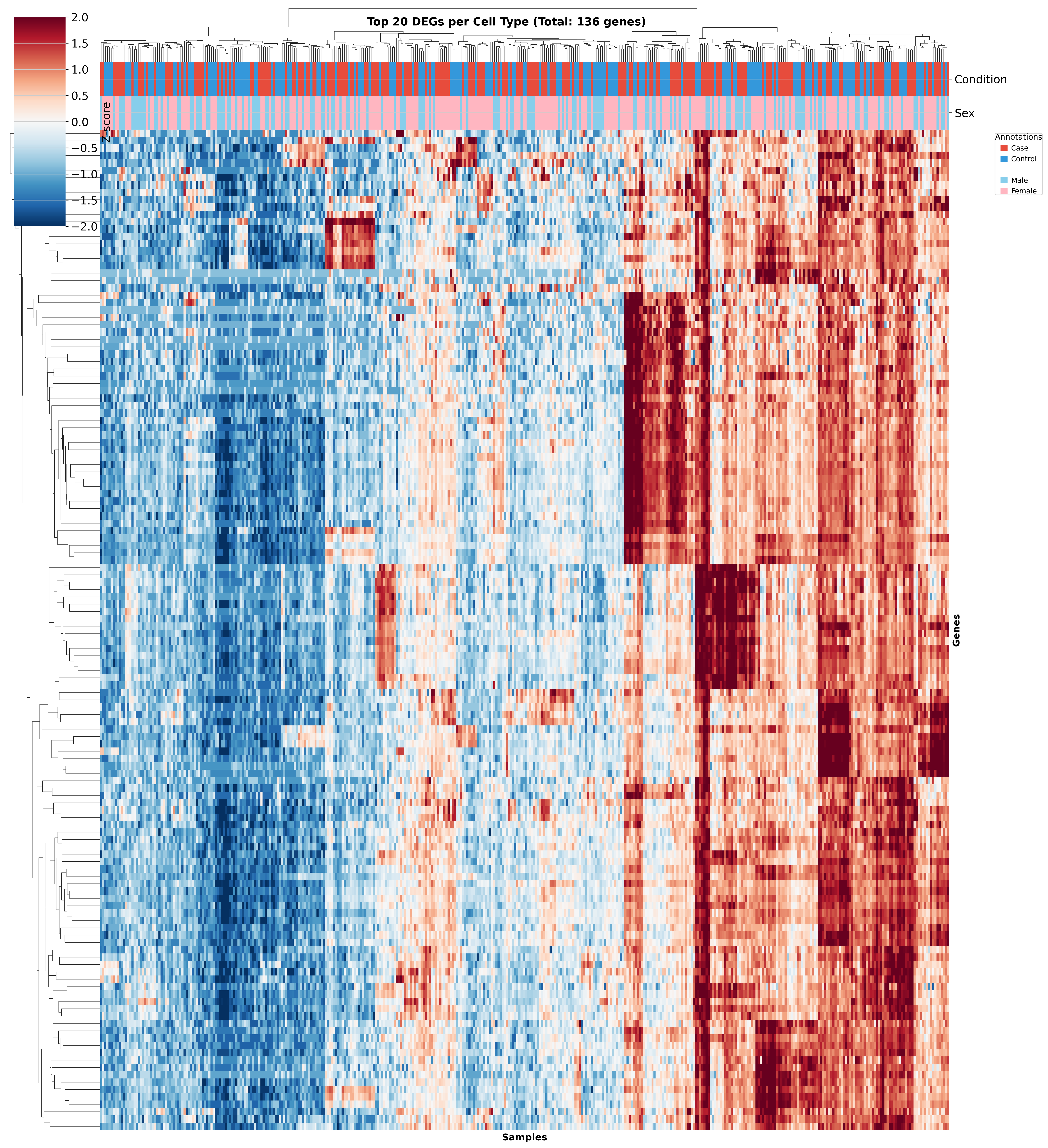
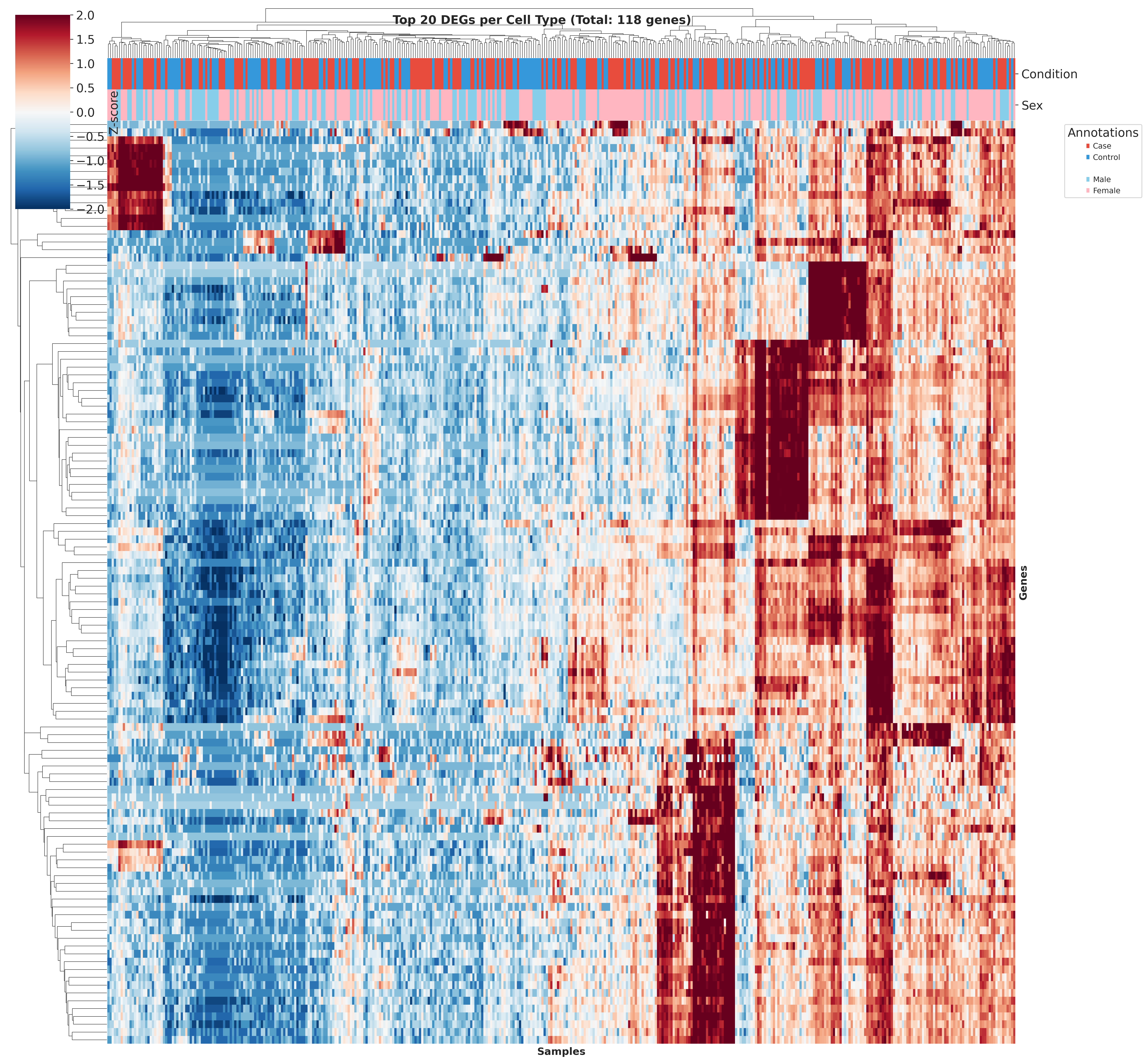
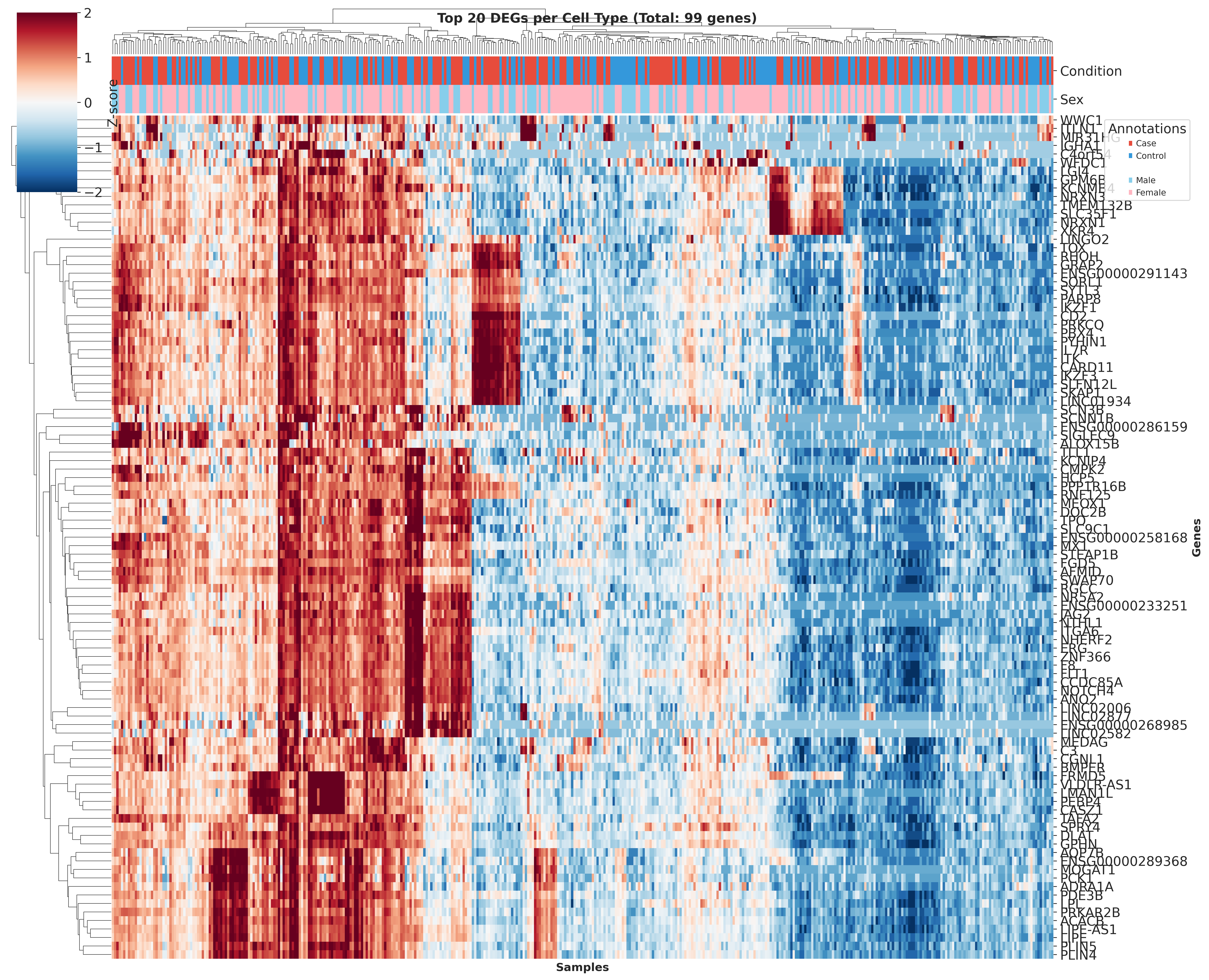
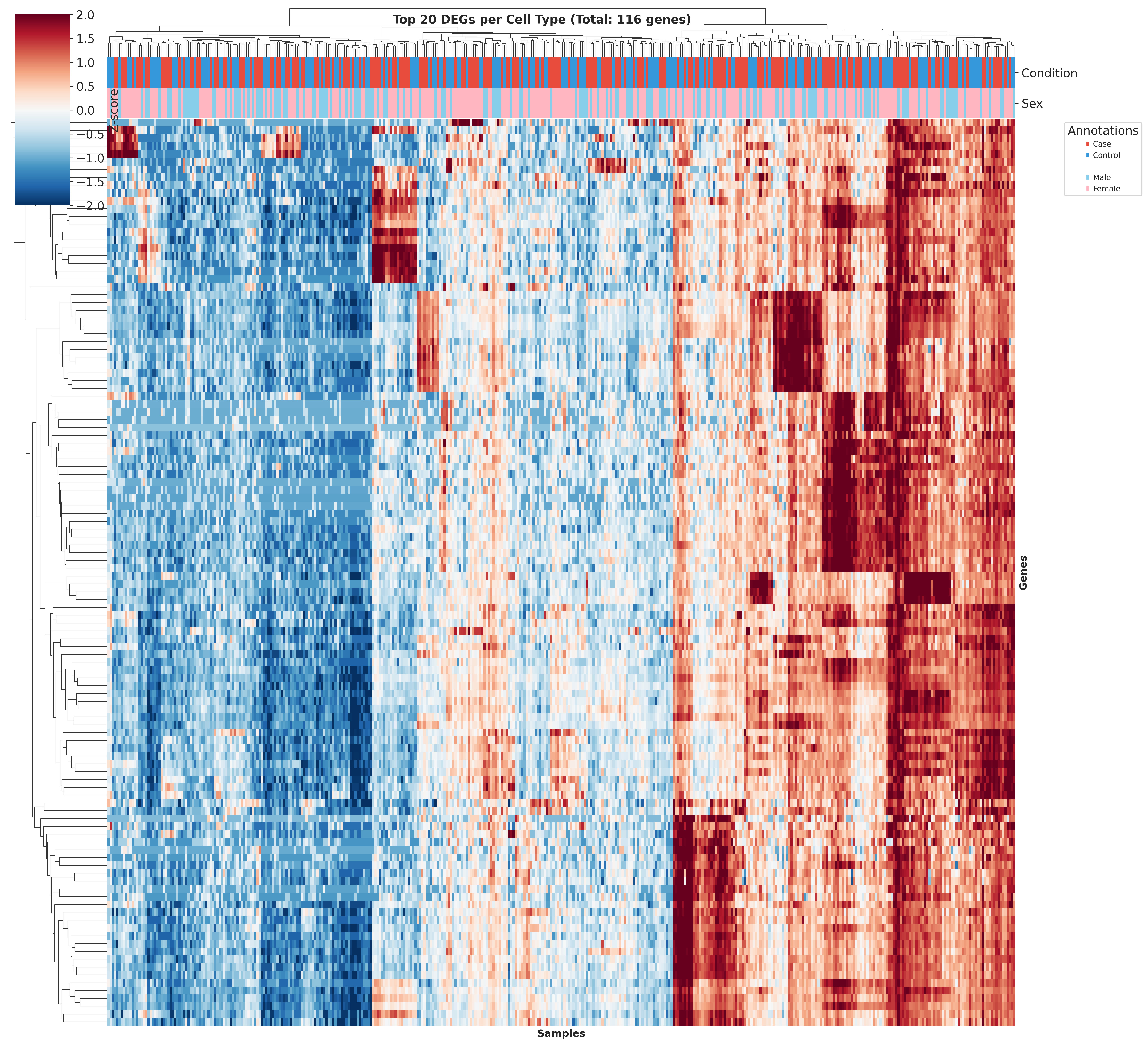
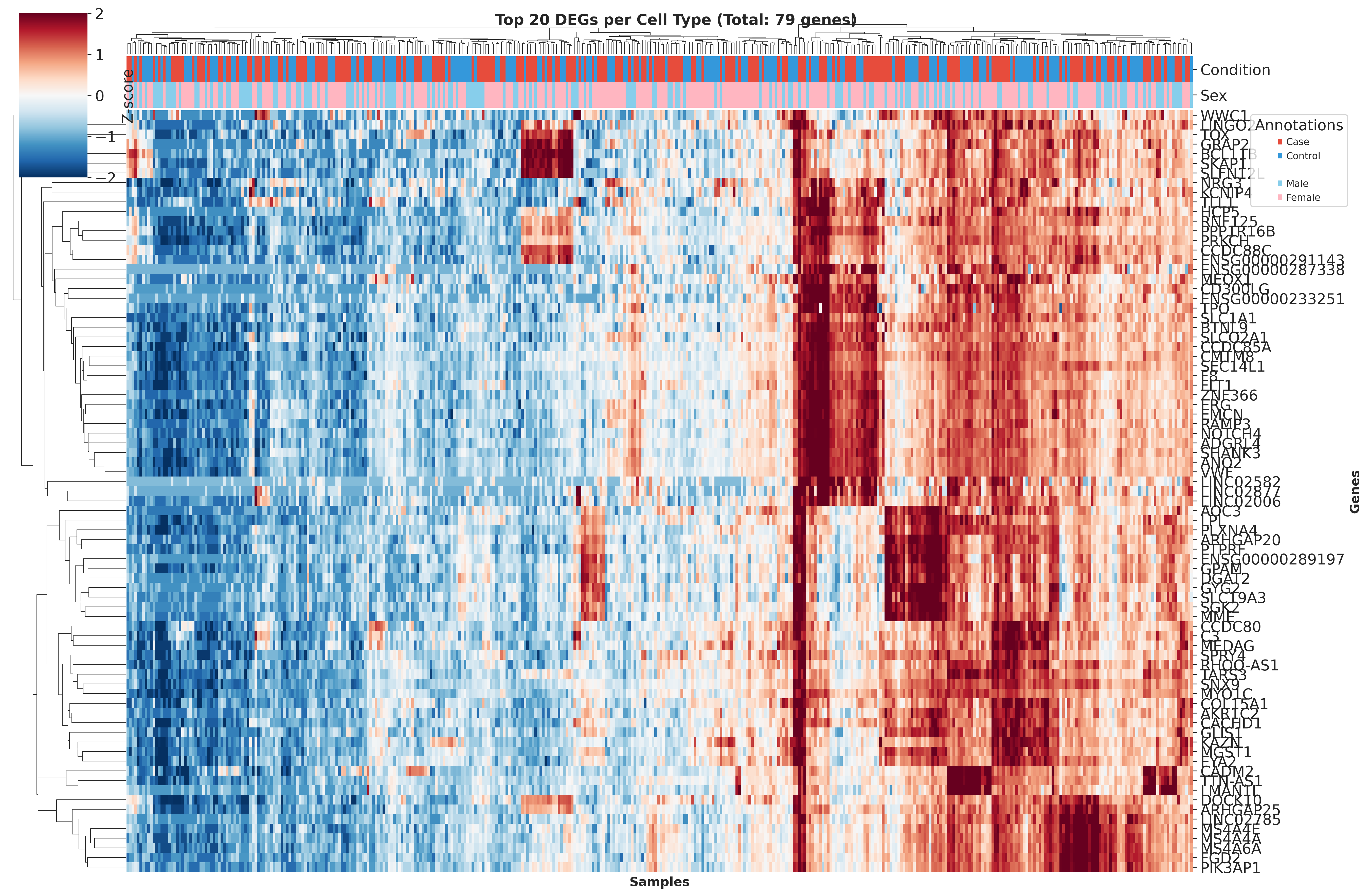
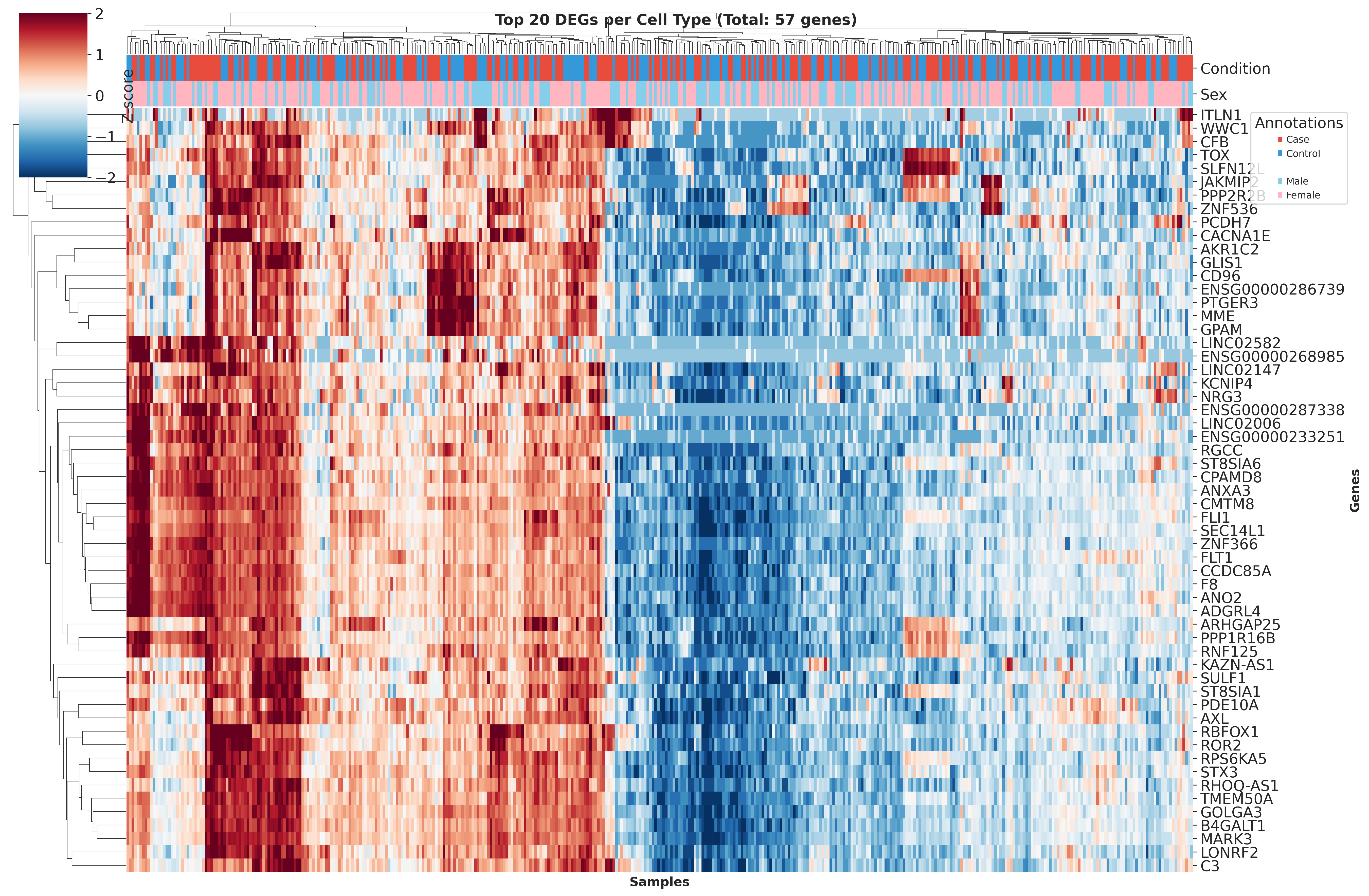
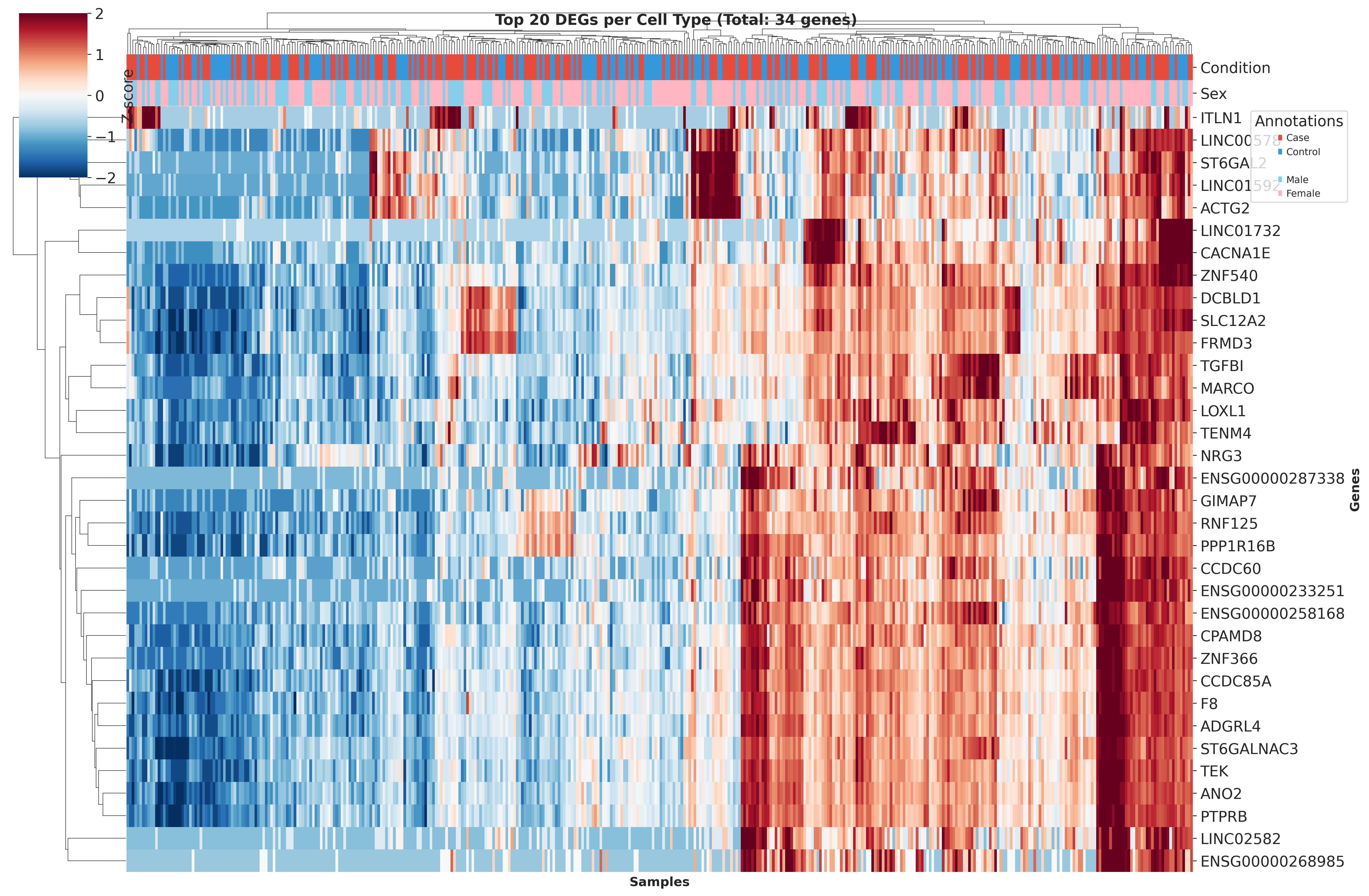
These heatmaps visualize the top DEGs identified in each cell type across different QC strategies. While core gene signatures remain similar, the ranking and significance of individual genes shift with different QC approaches.
Conclusion
This comprehensive comparison reveals several key insights:
- QC methods filter different cells: Each approach targets distinct subpopulations, with limited overlap between methods
- Clustering is relatively robust: Major cell populations remain identifiable regardless of QC choice
- Cell type annotation is consistent: The ability to identify known cell types is not dramatically affected by QC strategy
- DEG detection is sensitive to QC: Statistical significance of differential expression varies substantially with QC method, even when effect sizes are similar
The choice of QC method should be guided by the specific biological question and the tolerance for false positives versus false negatives in downstream analysis. For exploratory analysis, conservative QC may be preferable to retain biological diversity. For focused hypothesis testing, more stringent QC may improve signal-to-noise ratio.
References
- QClus: https://doi.org/10.1093/nar/gkae1145
- SoupX: https://doi.org/10.1093/gigascience/giaa151
- Scrublet: https://doi.org/10.1016/j.cmet.2018.11.005
- GEO Dataset GSE255612: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE255612
- Hill et al. (2024), PubMed ID 39562555: https://pubmed.ncbi.nlm.nih.gov/39562555/